dataset
Type of resources
Available actions
Topics
Keywords
Contact for the resource
Provided by
Years
Formats
Representation types
Update frequencies
status
Scale
Resolution
-
421: Vegetated low-lying areas in the coastal zone, above the high-tide line, susceptible to flooding by seawater. Often in the process of being filled in by coastal mud and sand sediments, gradually being colonized by halophilic plants. Salt marshes are in most cases directly connected to intertidal areas and may successively develop from them in the long-term. Salt-pans for extraction of salt from salt water by evaporation, active or in process of abandonment. Sections of salt marsh exploited for the production of salt, clearly distinguishable from the rest of the marsh by their parcellation and embankment systems. Coastal zone under tidal influence between open sea and land, which is flooded by sea water regularly twice a day in a ca. 12 hours cycle. Area between the average lowest and highest sea water level at low tide and high tide. Generally non-vegetated expanses of mud, sand or rock lying between high and low water marks. The seaward boundary of intertidal flats may underlay constant change in geographical extent due to littoral morphodynamics. Range of water level between low tide and high tide may vary between decimeters and several meters in height.
-
osm: Commercial building aggregated and rasterized from OSM polygons, first to 10m spatial resolution and after downsampled to 30m by spatial average.
-
Overview: 333: Areas with sparse vegetation, covering 10-50% of surface. Includes steppes, tundra, lichen heath, badlands, karstic areas and scattered high-altitude vegetation. Scattered vegetation is composed of herbaceous and/or ligneous and semi-ligneous species, the rest of area is naturally bare ground. Traceability (lineage): This dataset was produced with a machine learning framework with several input datasets, specified in detail in Witjes et al., 2022 (in review, preprint available at https://doi.org/10.21203/rs.3.rs-561383/v3 ) Scientific methodology: The single-class probability layers were generated with a spatiotemporal ensemble machine learning framework detailed in Witjes et al., 2022 (in review, preprint available at https://doi.org/10.21203/rs.3.rs-561383/v3 ). The single-class uncertainty layers were calculated by taking the standard deviation of the three single-class probabilities predicted by the three components of the ensemble. The HCL (hard class) layers represents the class with the highest probability as predicted by the ensemble. Usability: The HCL layers have a decreasing average accuracy (weighted F1-score) at each subsequent level in the CLC hierarchy. These metrics are 0.83 at level 1 (5 classes):, 0.63 at level 2 (14 classes), and 0.49 at level 3 (43 classes). This means that the hard-class maps are more reliable when aggregating classes to a higher level in the hierarchy (e.g. 'Discontinuous Urban Fabric' and 'Continuous Urban Fabric' to 'Urban Fabric'). Some single-class probabilities may more closely represent actual patterns for some classes that were overshadowed by unequal sample point distributions. Users are encouraged to set their own thresholds when postprocessing these datasets to optimize the accuracy for their specific use case. Uncertainty quantification: Uncertainty is quantified by taking the standard deviation of the probabilities predicted by the three components of the spatiotemporal ensemble model. Data validation approaches: The LULC classification was validated through spatial 5-fold cross-validation as detailed in the accompanying publication. Completeness: The dataset has chunks of empty predictions in regions with complex coast lines (e.g. the Zeeland province in the Netherlands and the Mar da Palha bay area in Portugal). These are artifacts that will be avoided in subsequent versions of the LULC product. Consistency: The accuracy of the predictions was compared per year and per 30km*30km tile across europe to derive temporal and spatial consistency by calculating the standard deviation. The standard deviation of annual weighted F1-score was 0.135, while the standard deviation of weighted F1-score per tile was 0.150. This means the dataset is more consistent through time than through space: Predictions are notably less accurate along the Mediterrranean coast. The accompanying publication contains additional information and visualisations. Positional accuracy: The raster layers have a resolution of 30m, identical to that of the Landsat data cube used as input features for the machine learning framework that predicted it. Temporal accuracy: The dataset contains predictions and uncertainty layers for each year between 2000 and 2019. Thematic accuracy: The maps reproduce the Corine Land Cover classification system, a hierarchical legend that consists of 5 classes at the highest level, 14 classes at the second level, and 44 classes at the third level. Class 523: Oceans was omitted due to computational constraints.
-
Overview: Actual Natural Vegetation (ANV): probability of occurrence for the Common hazel in its realized environment for the period 2000 - 2022 Traceability (lineage): This is an original dataset produced with a machine learning framework which used a combination of point datasets and raster datasets as inputs. Point dataset is a harmonized collection of tree occurrence data, comprising observations from National Forest Inventories (EU-Forest), GBIF and LUCAS. The complete dataset is available on Zenodo. Raster datasets used as input are: harmonized and gapfilled time series of seasonal aggregates of the Landsat GLAD ARD dataset (bands and spectral indices); monthly time series air and surface temperature and precipitation from a reprocessed version of the Copernicus ERA5 dataset; long term averages of bioclimatic variables from CHELSA, tree species distribution maps from the European Atlas of Forest Tree Species; elevation, slope and other elevation-derived metrics; long term monthly averages snow probability and long term monthly averages of cloud fraction from MODIS. For a more comprehensive list refer to Bonannella et al. (2022) (in review, preprint available at: https://doi.org/10.21203/rs.3.rs-1252972/v1). Scientific methodology: Probability and uncertainty maps were the output of a spatiotemporal ensemble machine learning framework based on stacked regularization. Three base models (random forest, gradient boosted trees and generalized linear models) were first trained on the input dataset and their predictions were used to train an additional model (logistic regression) which provided the final predictions. More details on the whole workflow are available in the listed publication. Usability: Probability maps can be used to detect potential forest degradation and compositional change across the time period analyzed. Some possible applications for these topics are explained in the listed publication. Uncertainty quantification: Uncertainty is quantified by taking the standard deviation of the probabilities predicted by the three components of the spatiotemporal ensemble model. Data validation approaches: Distribution maps were validated using a spatial 5-fold cross validation following the workflow detailed in the listed publication. Completeness: The raster files perfectly cover the entire Geo-harmonizer region as defined by the landmask raster dataset available here. Consistency: Areas which are outside of the calibration area of the point dataset (Iceland, Norway) usually have high uncertainty values. This is not only a problem of extrapolation but also of poor representation in the feature space available to the model of the conditions that are present in this countries. Positional accuracy: The rasters have a spatial resolution of 30m. Temporal accuracy: The maps cover the period 2000 - 2020, each map covers a certain number of years according to the following scheme: (1) 2000--2002, (2) 2002--2006, (3) 2006--2010, (4) 2010--2014, (5) 2014--2018 and (6) 2018--2020 Thematic accuracy: Both probability and uncertainty maps contain values from 0 to 100: in the case of probability maps, they indicate the probability of occurrence of a single individual of the target species, while uncertainty maps indicate the standard deviation of the ensemble model.
-
Overview: Actual Natural Vegetation (ANV): probability of occurrence for the Austrian pine in its realized environment for the period 2000 - 2027 Traceability (lineage): This is an original dataset produced with a machine learning framework which used a combination of point datasets and raster datasets as inputs. Point dataset is a harmonized collection of tree occurrence data, comprising observations from National Forest Inventories (EU-Forest), GBIF and LUCAS. The complete dataset is available on Zenodo. Raster datasets used as input are: harmonized and gapfilled time series of seasonal aggregates of the Landsat GLAD ARD dataset (bands and spectral indices); monthly time series air and surface temperature and precipitation from a reprocessed version of the Copernicus ERA5 dataset; long term averages of bioclimatic variables from CHELSA, tree species distribution maps from the European Atlas of Forest Tree Species; elevation, slope and other elevation-derived metrics; long term monthly averages snow probability and long term monthly averages of cloud fraction from MODIS. For a more comprehensive list refer to Bonannella et al. (2022) (in review, preprint available at: https://doi.org/10.21203/rs.3.rs-1252972/v1). Scientific methodology: Probability and uncertainty maps were the output of a spatiotemporal ensemble machine learning framework based on stacked regularization. Three base models (random forest, gradient boosted trees and generalized linear models) were first trained on the input dataset and their predictions were used to train an additional model (logistic regression) which provided the final predictions. More details on the whole workflow are available in the listed publication. Usability: Probability maps can be used to detect potential forest degradation and compositional change across the time period analyzed. Some possible applications for these topics are explained in the listed publication. Uncertainty quantification: Uncertainty is quantified by taking the standard deviation of the probabilities predicted by the three components of the spatiotemporal ensemble model. Data validation approaches: Distribution maps were validated using a spatial 5-fold cross validation following the workflow detailed in the listed publication. Completeness: The raster files perfectly cover the entire Geo-harmonizer region as defined by the landmask raster dataset available here. Consistency: Areas which are outside of the calibration area of the point dataset (Iceland, Norway) usually have high uncertainty values. This is not only a problem of extrapolation but also of poor representation in the feature space available to the model of the conditions that are present in this countries. Positional accuracy: The rasters have a spatial resolution of 30m. Temporal accuracy: The maps cover the period 2000 - 2020, each map covers a certain number of years according to the following scheme: (1) 2000--2002, (2) 2002--2006, (3) 2006--2010, (4) 2010--2014, (5) 2014--2018 and (6) 2018--2020 Thematic accuracy: Both probability and uncertainty maps contain values from 0 to 100: in the case of probability maps, they indicate the probability of occurrence of a single individual of the target species, while uncertainty maps indicate the standard deviation of the ensemble model.
-
Overview: Actual Natural Vegetation (ANV): probability of occurrence for the Turkey oak in its realized environment for the period 2000 - 2031 Traceability (lineage): This is an original dataset produced with a machine learning framework which used a combination of point datasets and raster datasets as inputs. Point dataset is a harmonized collection of tree occurrence data, comprising observations from National Forest Inventories (EU-Forest), GBIF and LUCAS. The complete dataset is available on Zenodo. Raster datasets used as input are: harmonized and gapfilled time series of seasonal aggregates of the Landsat GLAD ARD dataset (bands and spectral indices); monthly time series air and surface temperature and precipitation from a reprocessed version of the Copernicus ERA5 dataset; long term averages of bioclimatic variables from CHELSA, tree species distribution maps from the European Atlas of Forest Tree Species; elevation, slope and other elevation-derived metrics; long term monthly averages snow probability and long term monthly averages of cloud fraction from MODIS. For a more comprehensive list refer to Bonannella et al. (2022) (in review, preprint available at: https://doi.org/10.21203/rs.3.rs-1252972/v1). Scientific methodology: Probability and uncertainty maps were the output of a spatiotemporal ensemble machine learning framework based on stacked regularization. Three base models (random forest, gradient boosted trees and generalized linear models) were first trained on the input dataset and their predictions were used to train an additional model (logistic regression) which provided the final predictions. More details on the whole workflow are available in the listed publication. Usability: Probability maps can be used to detect potential forest degradation and compositional change across the time period analyzed. Some possible applications for these topics are explained in the listed publication. Uncertainty quantification: Uncertainty is quantified by taking the standard deviation of the probabilities predicted by the three components of the spatiotemporal ensemble model. Data validation approaches: Distribution maps were validated using a spatial 5-fold cross validation following the workflow detailed in the listed publication. Completeness: The raster files perfectly cover the entire Geo-harmonizer region as defined by the landmask raster dataset available here. Consistency: Areas which are outside of the calibration area of the point dataset (Iceland, Norway) usually have high uncertainty values. This is not only a problem of extrapolation but also of poor representation in the feature space available to the model of the conditions that are present in this countries. Positional accuracy: The rasters have a spatial resolution of 30m. Temporal accuracy: The maps cover the period 2000 - 2020, each map covers a certain number of years according to the following scheme: (1) 2000--2002, (2) 2002--2006, (3) 2006--2010, (4) 2010--2014, (5) 2014--2018 and (6) 2018--2020 Thematic accuracy: Both probability and uncertainty maps contain values from 0 to 100: in the case of probability maps, they indicate the probability of occurrence of a single individual of the target species, while uncertainty maps indicate the standard deviation of the ensemble model.
-
Overview: 313: Vegetation formation composed principally of trees, including shrub and bush understory,where neither broad-leaved nor coniferous species predominate. Traceability (lineage): This dataset was produced with a machine learning framework with several input datasets, specified in detail in Witjes et al., 2022 (in review, preprint available at https://doi.org/10.21203/rs.3.rs-561383/v3 ) Scientific methodology: The single-class probability layers were generated with a spatiotemporal ensemble machine learning framework detailed in Witjes et al., 2022 (in review, preprint available at https://doi.org/10.21203/rs.3.rs-561383/v3 ). The single-class uncertainty layers were calculated by taking the standard deviation of the three single-class probabilities predicted by the three components of the ensemble. The HCL (hard class) layers represents the class with the highest probability as predicted by the ensemble. Usability: The HCL layers have a decreasing average accuracy (weighted F1-score) at each subsequent level in the CLC hierarchy. These metrics are 0.83 at level 1 (5 classes):, 0.63 at level 2 (14 classes), and 0.49 at level 3 (43 classes). This means that the hard-class maps are more reliable when aggregating classes to a higher level in the hierarchy (e.g. 'Discontinuous Urban Fabric' and 'Continuous Urban Fabric' to 'Urban Fabric'). Some single-class probabilities may more closely represent actual patterns for some classes that were overshadowed by unequal sample point distributions. Users are encouraged to set their own thresholds when postprocessing these datasets to optimize the accuracy for their specific use case. Uncertainty quantification: Uncertainty is quantified by taking the standard deviation of the probabilities predicted by the three components of the spatiotemporal ensemble model. Data validation approaches: The LULC classification was validated through spatial 5-fold cross-validation as detailed in the accompanying publication. Completeness: The dataset has chunks of empty predictions in regions with complex coast lines (e.g. the Zeeland province in the Netherlands and the Mar da Palha bay area in Portugal). These are artifacts that will be avoided in subsequent versions of the LULC product. Consistency: The accuracy of the predictions was compared per year and per 30km*30km tile across europe to derive temporal and spatial consistency by calculating the standard deviation. The standard deviation of annual weighted F1-score was 0.135, while the standard deviation of weighted F1-score per tile was 0.150. This means the dataset is more consistent through time than through space: Predictions are notably less accurate along the Mediterrranean coast. The accompanying publication contains additional information and visualisations. Positional accuracy: The raster layers have a resolution of 30m, identical to that of the Landsat data cube used as input features for the machine learning framework that predicted it. Temporal accuracy: The dataset contains predictions and uncertainty layers for each year between 2000 and 2019. Thematic accuracy: The maps reproduce the Corine Land Cover classification system, a hierarchical legend that consists of 5 classes at the highest level, 14 classes at the second level, and 44 classes at the third level. Class 523: Oceans was omitted due to computational constraints.
-
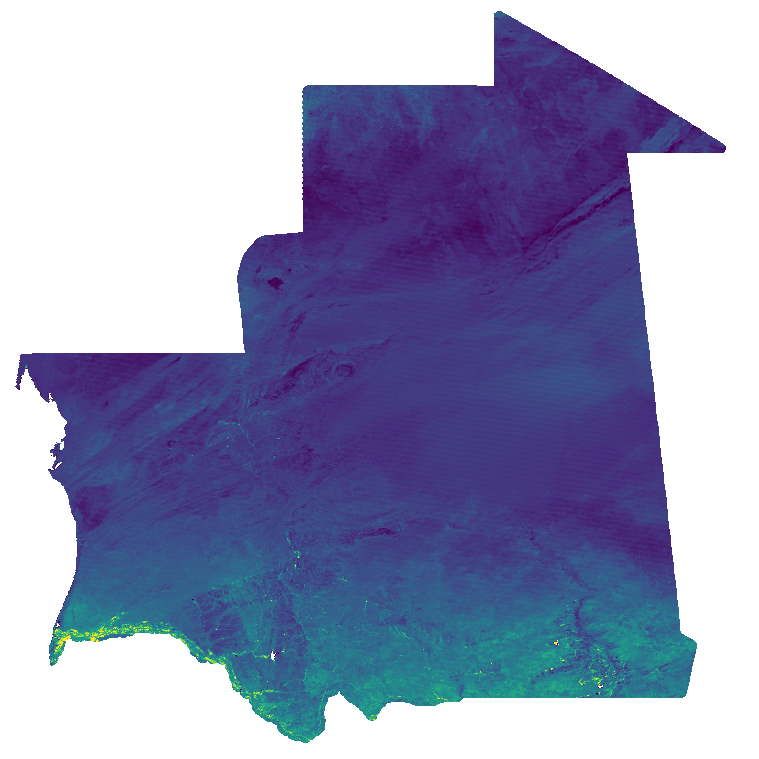
Normalized Difference Vegetation Index (NDVI) from MODIS data for Mauritania at 30 arc seconds (ca. 1000 meter) resolution (2019 - 2023). Source data: - MODIS/Terra Vegetation Indices 16-Day L3 Global 1 km SIN Grid (MOD13A2 v061): https://lpdaac.usgs.gov/products/mod13a2v061/ The Terra Moderate Resolution Imaging Spectroradiometer (MODIS) Vegetation Indices 16-Day (MOD13A2) Version 6.1 product provides Vegetation Index (VI) values at a per pixel basis at 1 kilometer (km) spatial resolution. There are two primary vegetation layers. The first is the Normalized Difference Vegetation Index (NDVI), which is referred to as the continuity index to the existing National Oceanic and Atmospheric Administration-Advanced Very High Resolution Radiometer (NOAA-AVHRR) derived NDVI. The second vegetation layer is the Enhanced Vegetation Index (EVI), which has improved sensitivity over high biomass regions. The algorithm for this product chooses the best available pixel value from all the acquisitions from the 16 day period. The criteria used is low clouds, low view angle and the highest NDVI/EVI value. For the time period January 2019 - December 2023, the NDVI layer of the original data has been processed. Bad quality pixels or pixels with snow/ice and/or cloud cover have been masked using the provided quality assurance (QA) layers and appear as "no data". These 16-Day data are then aggregated to monthly temporal resolution using the maximum and reprojected to Latitude-Longitude/WGS84. File naming: ndvi_filt_YYYY_MM_01T00_00_00.tif e.g.: ndvi_filt_2023_12_01T00_00_00.tif The date within the filename is year and month of aggregated timestamp. Pixel values: NDVI * 10000 Scaled to Integer, example: value 6473 = 0.6473 Projection + EPSG code: Latitude-Longitude/WGS84 (EPSG: 4326) Spatial extent: north: 28N south: 14N west: 18W east: 4W Temporal extent: January 2019 - December 2023 Spatial resolution: 30 arc seconds (approx. 1000 m) Temporal resolution: monthly Software used: GRASS GIS 8.3.2 Format: GeoTIFF Original dataset license: All data products distributed by NASA's Land Processes Distributed Active Archive Center (LP DAAC) are available at no charge. The LP DAAC requests that any author using NASA data products in their work provide credit for the data, and any assistance provided by the LP DAAC, in the data section of the paper, the acknowledgement section, and/or as a reference. The recommended citation for each data product is available on its Digital Object Identifier (DOI) Landing page, which can be accessed through the Search Data Catalog interface. For more information see: https://lpdaac.usgs.gov/products/mod13a2v061/ Processed by: mundialis GmbH & Co. KG, Germany (https://www.mundialis.de/) Contact: mundialis GmbH & Co. KG, info@mundialis.de Acknowledgements: This study was partially funded by EU grant 874850 MOOD. The contents of this publication are the sole responsibility of the authors and don't necessarily reflect the views of the European Commission.
-
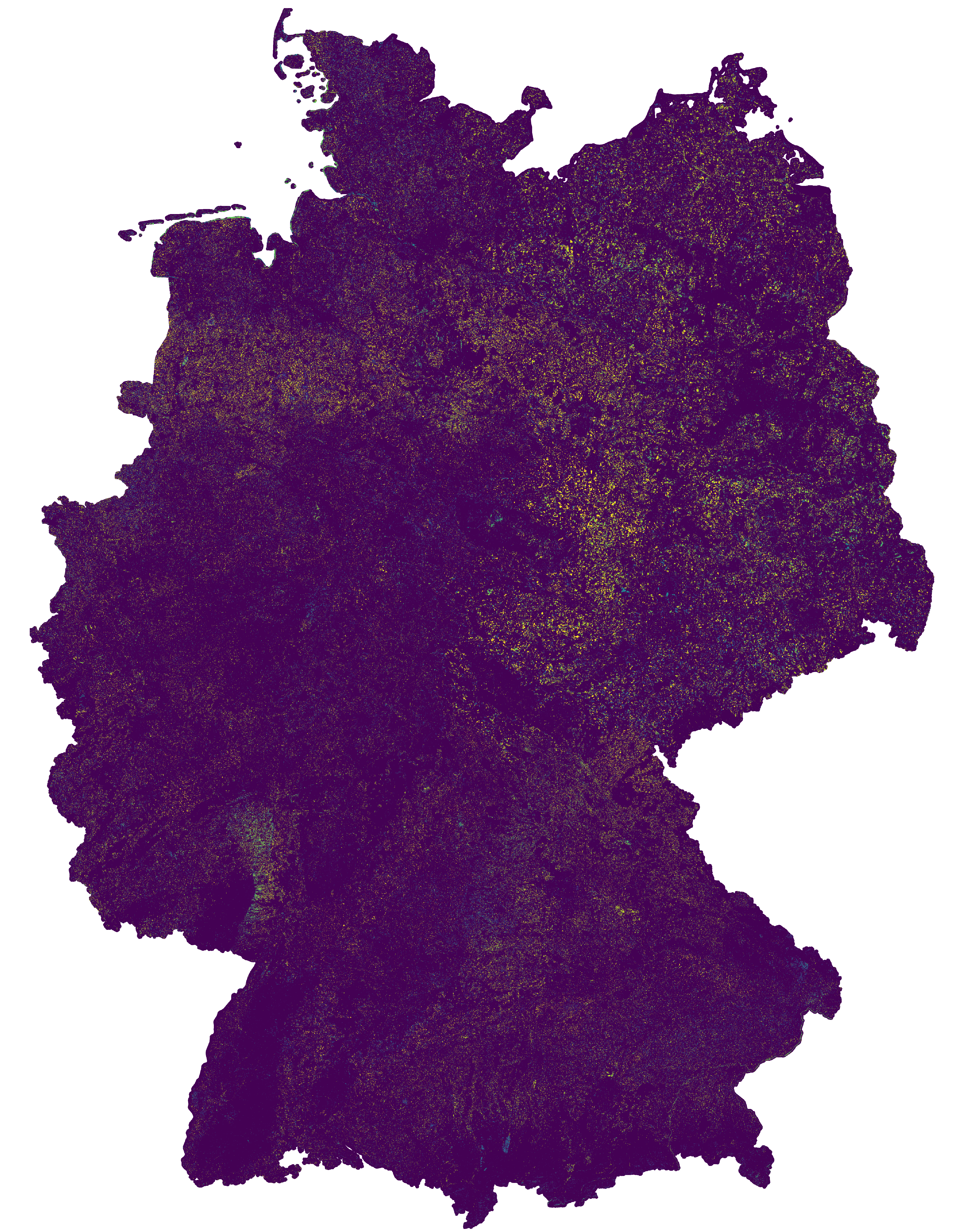
This change map was produced as an intermediate result in the course of the project incora (Inwertsetzung von Copernicus-Daten für die Raumbeobachtung, mFUND Förderkennzeichen: 19F2079C) in cooperation with ILS (Institut für Landes- und Stadtentwicklungsforschung gGmbH) and BBSR (Bundesinstitut für Bau-, Stadt- und Raumforschung) funded by BMVI (Federal Ministry of Transport and Digital Infrastructure). The goal of incora is an analysis of settlement and infrastructure dynamics in Germany based on Copernicus Sentinel data. The map indicates land cover changes between the years 2016 and 2019. It is a difference map from two classifications based on Sentinel-2 MAJA data (MAJA L3A-WASP: https://geoservice.dlr.de/web/maps/sentinel2:l3a:wasp; DLR (2019): Sentinel-2 MSI - Level 2A (MAJA-Tiles)- Germany). More information on the two basis classifications can be found here: https://data.mundialis.de/geonetwork/srv/eng/catalog.search#/metadata/db130a09-fc2e-421d-95e2-1575e7c4b45c https://data.mundialis.de/geonetwork/srv/eng/catalog.search#/metadata/36512b46-f3aa-4aa4-8281-7584ec46c813 To keep only significant changes in the change detection map, the following postprocessing steps are applied to the initial difference raster: - Modefilter (3x3) to eliminate isolated pixels and edge effects - Information gain in a 4x4 window compares class distribution within the window from the two timesteps. High values indicate that the class distribution in the window has changed, and thus a change is likely. Gain ranges from 0 to 1, all changes < 0.5 are omitted. - Change areas < 1ha are removed The resulting map has the following nomenclature: 0: No Change 1: Change from low vegetation to forest 2: Change from water to forest 3: Change from built-up to forest 4: Change from bare soil to forest 5: Change from agriculture to forest 6: Change from forest to low vegetation 7: Change from water to low vegetation 8: Change from built-up to low vegetation 9: Change from bare soil to low vegetation 10: Change from agriculture to low vegetation 11: Change from forest to water 12: Change from low vegetation to water 13: Change from built-up to water 14: Change from bare soil to water 15: Change from agriculture to water 16: Change from forest to built-up 17: Change from low vegetation to built-up 18: Change from water to built-up 19: Change from bare soil to built-up 20: Change from agriculture to built-up 21: Change from forest to bare soil 22: Change from low vegetation to bare soil 23: Change from water to bare soil 24: Change from built-up to bare soil 25: Change from agriculture to bare soil 26: Change from forest to agriculture 27: Change from low vegetation to agriculture 28: Change from water to agriculture 29: Change from built-up to agriculture 30: Change from bare soil to agriculture - Contains modified Copernicus Sentinel data (2016/2019), processed by mundialis Incora report with details on methods and results: pending
-
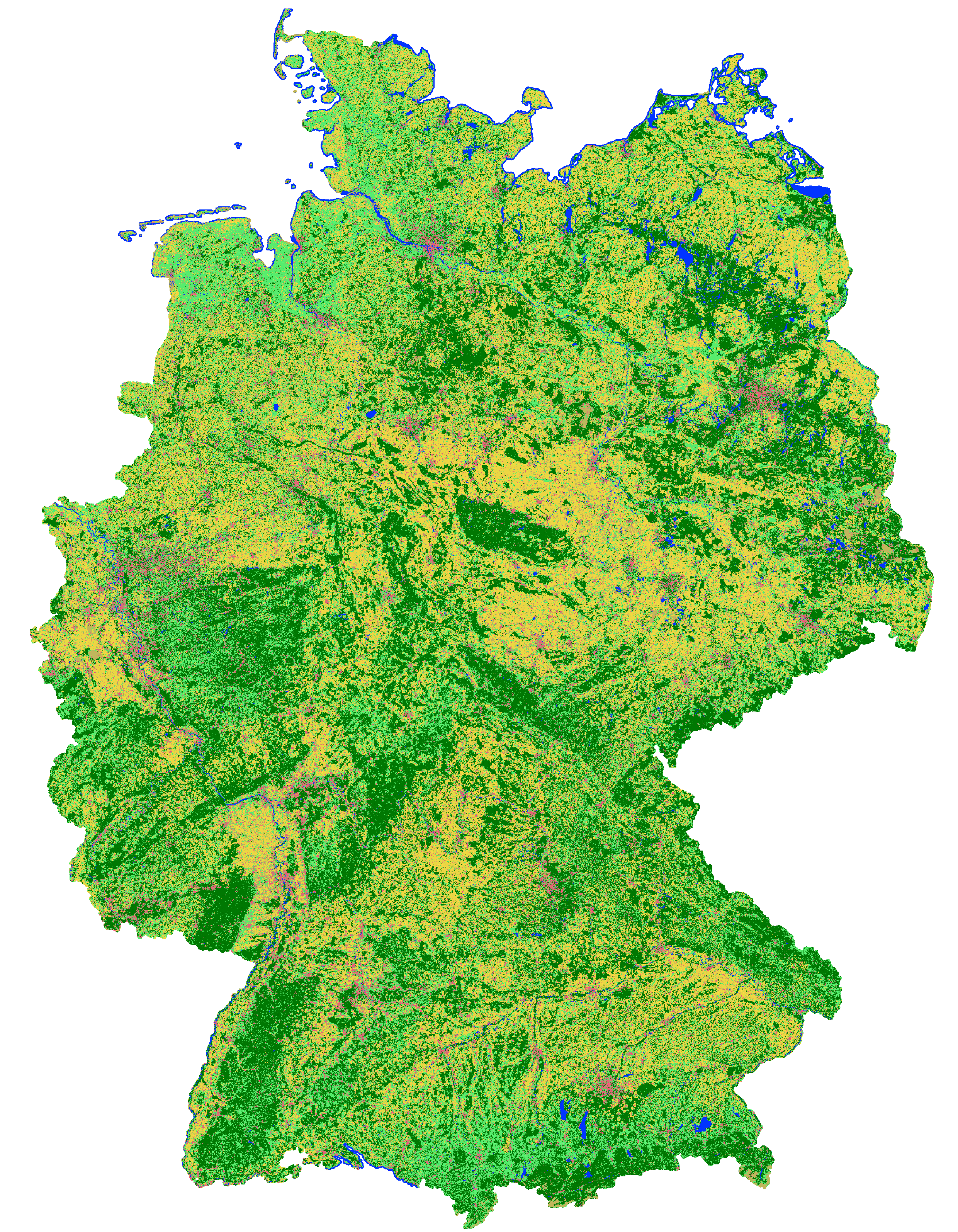
This landcover map was produced as an intermediate result in the course of the project incora (Inwertsetzung von Copernicus-Daten für die Raumbeobachtung, mFUND Förderkennzeichen: 19F2079C) in cooperation with ILS (Institut für Landes- und Stadtentwicklungsforschung gGmbH) and BBSR (Bundesinstitut für Bau-, Stadt- und Raumforschung) funded by BMVI (Federal Ministry of Transport and Digital Infrastructure). The goal of incora is an analysis of settlement and infrastructure dynamics in Germany based on Copernicus Sentinel data. This classification is based on a time-series of monthly averaged, atmospherically corrected Sentinel-2 tiles (MAJA L3A-WASP: https://geoservice.dlr.de/web/maps/sentinel2:l3a:wasp; DLR (2019): Sentinel-2 MSI - Level 2A (MAJA-Tiles)- Germany). It consists of the following landcover classes: 10: forest 20: low vegetation 30: water 40: built-up 50: bare soil 60: agriculture Potential training and validation areas were automatically extracted using spectral indices and their temporal variability from the Sentinel-2 data itself as well as the following auxiliary datasets: - OpenStreetMap (Map data copyrighted OpenStreetMap contributors and available from htttps://www.openstreetmap.org) - Copernicus HRL Imperviousness Status Map 2018 (© European Union, Copernicus Land Monitoring Service 2018, European Environment Agency (EEA)) - S2GLC Land Cover Map of Europe 2017 (Malinowski et al. 2020: Automated Production of Land Cover/Use Map of Europe Based on Sentinel-2 Imagery. Remote Sens. 2020, 12(21), 3523; https://doi.org/10.3390/rs12213523) - Germany NUTS administrative areas 1:250000 (© GeoBasis-DE / BKG 2020 / dl-de/by-2-0 / https://gdz.bkg.bund.de/index.php/default/nuts-gebiete-1-250-000-stand-31-12-nuts250-31-12.html) - Contains modified Copernicus Sentinel data (2016), processed by mundialis Processing was performed for blocks of federal states and individual maps were mosaicked afterwards. For each class 100,000 pixels from the potential training areas were extracted as training data. An exemplary validation of the classification results was perfomed for the federal state of North Rhine-Westphalia as its open data policy allows for direct access to official data to be used as reference. Rules to convert relevant ATKIS Basis-DLM object classes to the incora nomenclature were defined. Subsequently, 5.000 reference points were randomly sampled and their classification in each case visually examined and, if necessary, revised to obtain a robust reference data set. The comparison of this reference data set with the incora classification yielded the following results: overall accurary: 88.4% class: user's accuracy / producer's accurary (number of reference points n) forest: 96.7% / 94.3% (1410) low vegetation: 70.6% / 84.0% (844) water: 98.5% / 94.2% (69) built-up: 98.2% / 89.8% (983) bare soil: 19.7% / 58.5% (41) agriculture: 91.7% / 85.3% (1653) Incora report with details on methods and results: pending
 Open Data Science Europe Metadata Catalog
Open Data Science Europe Metadata Catalog